download.file("https://ndownloader.figshare.com/files/2292169",
"data/portal_data_joined.csv")Week 3 - Working with data
What is tabular data?
What it is
Rows = observations, columns = variables, cells = individual observations (Broman and Woo 2018)
Spreadsheets and relational databases
What it isn’t
Geospatial data (vectors and rasters)
Phylogenetic data (trees)
What is the tidyverse?
“At a high level, the tidyverse is a language for solving data science challenges with R code. Its primary goal is to facilitate a conversation between a human and a computer about data. Less abstractly, the tidyverse is a collection of R packages that share a high-level design philosophy and low-level grammar and data structures, so that learning one package makes it easier to learn the next.” (Wickham et al. 2019)
Core packages attached on library(tidyverse)
dplyrreadrtidyrggplot2and several others
Other installed packages on install.packages("tidyverse")
lubridatereadxlgoogledriveand several others
So this is redundant:
library(tidyverse)
library(dplyr)
library(ggplot2)But this isn’t:
library(tidyverse)
library(lubridate)Core principles
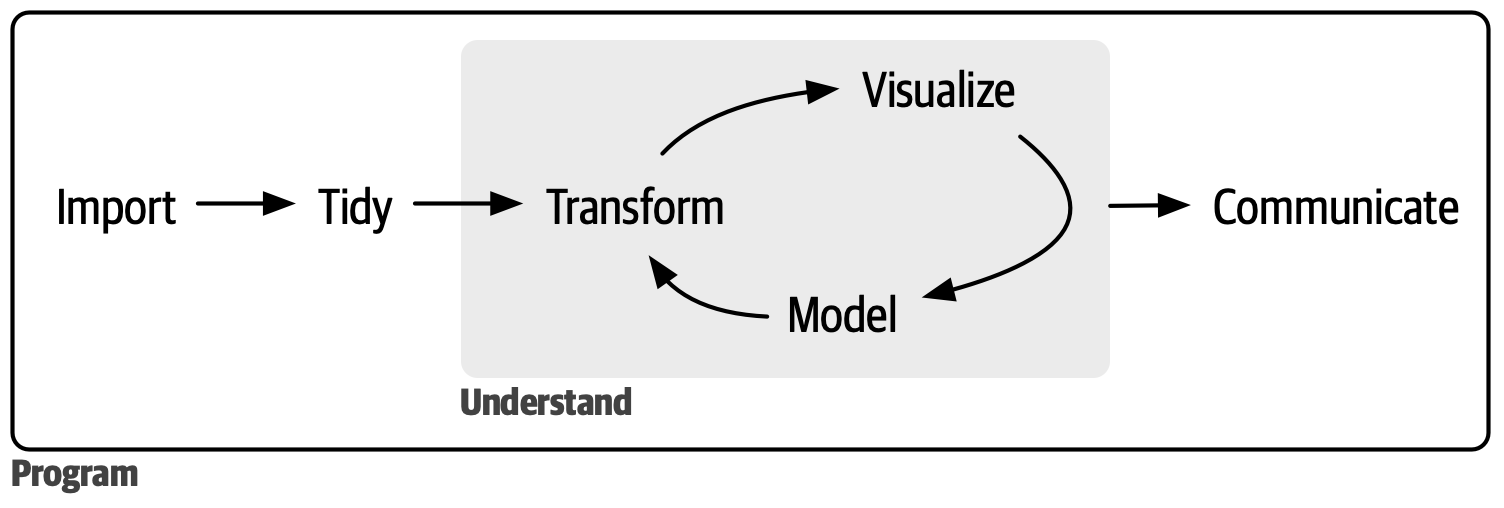
Use similar patterns and conventions for every step of the data science workflow
Emphasis on data frames, the ubiquitous data structure in R for tabular data
Tidy data. Rows = observations, columns = variables, cells = individual observations (Broman and Woo 2018)
%>%Pipe friendly! More on that shortly.
Project set up
- Create a project in RStudio called
bioe215_working_data. Make sure it has a git repo. - Create a GitHub repo to go with it.
- Create folders for
data,scratch,reports, anddocs. - Download the sample data.
- Create an R script
scratch/lesson3.R.
Data transformations with dplyr
Follow along in scratch/lesson3.R.
First things first: library(tidyverse)
Load the data
Read and inspect
surveys <- read_csv("data/portal_data_joined.csv")
head(surveys)
summary(surveys)Q1: What’s the type of column species_id? Of hindfoot_length?
Q2: How many rows and columns are in surveys?
selecting columns and filtering rows
Many tidyverse functions let you treat columns in a data frame as variables. In the following sections, notice we’re not putting column names in quotes.
How do I keep a few columns?
select(surveys, plot_id, species_id, weight)
select(surveys, plot_id, species_id, weight_g = weight)How do I remove a few columns?
select(surveys, -record_id, -species_id)How do I keep rows based on a condition?
filter(surveys, year == 1995)
filter(surveys, year == 1995, plot_id == 7)
filter(surveys, month == 2 | day == 20)Q3: filter() surveys to records collected in November where hindfoot_length is greater than 36.0
Q4: Fix these errors
filter(surveys, year = 1995)
filter(surveys, polt_id == 2)Pipes %>%
For a sequence of transformations, use pipes for readability
# Filter then select without pipes, two ways
# First way
select(filter(surveys, year == 1995), plot_id, species_id, weight)
# Second way
surveys_1995 <- filter(surveys, year == 1995)
surveys_psw <- select(surveys_1995, plot_id, species_id, weight)Instead, use pipes! Read them aloud as “then”.
surveys_psw <- surveys %>%
filter(year == 1995) %>%
select(plot_id, species_id, weight)Under the hood, the left hand side of the pipe becomes the first argument on the right hand side.
Q5: Use pipes to subset surveys to animals collected before 1995 retaining just the columns year, sex, and weight
Add/change columns with mutate
Add a column
surveys %>%
mutate(weight_kg = weight / 1000)Add multiple columns based on each other
surveys %>%
mutate(weight_kg = weight / 1000,
weight_lb = weight_kg * 2.2)So many NAs! Add a filter before mutate.
surveys %>%
filter(!is.na(weight)) %>%
mutate(weight_kg = weight / 1000,
weight_lb = weight_kg * 2.2)Convert year, month, and day to dates
surveys %>%
select(year, month, day) %>%
mutate(date_str = paste(year, month, day, sep = "-"),
date = as.Date(date_str))Q6: Create a new data frame from the surveys data that meets the following criteria: contains only the species_id column and a new column called hindfoot_cm containing the hindfoot_length values (currently in mm) converted to centimeters. In this hindfoot_cm column, there are no NAs and all values are less than 3.
Hint: think about how the commands should be ordered to produce this data frame!
Split-apply-combine with summarize
What’s the average weight of the observed animals by sex?
surveys %>%
group_by(sex) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE))Group by multiple columns, e.g. sex and species.
surveys %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE))Notice the warning message: our output is still grouped by species_id. By default, summarize() only removes one level of grouping. This usually leads to unexpected results. As the warning suggests, use .groups to drop all groups.
surveys %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE),
.groups = "drop")NaN is the result of calling mean() on an empty vector. Let’s remove NAs before summarizing.
surveys %>%
filter(!is.na(weight)) %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight),
.groups = "drop")Can also generate multiple summaries per group.
surveys %>%
filter(!is.na(weight)) %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight),
.groups = "drop")Sort with arrange
surveys %>%
filter(!is.na(weight)) %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight),
.groups = "drop") %>%
arrange(desc(mean_weight))Utility functions
count(), drop_na()
count() is a shortcut to getting the size of groups
surveys %>%
count(sex, species) %>%
arrange(species, desc(n))drop_na() is a shortcut for removing rows with missing values
surveys %>%
drop_na(weight, sex) %>%
group_by(species_id, sex) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight),
.groups = "drop") %>%
arrange(desc(mean_weight))Q7: How many animals were caught in each plot_type surveyed?
Q8: Use group_by() and summarize() to find the mean, min, and max hindfoot length for each species (using species_id). Also add the number of observations (hint: see ?n).
Q9: What was the heaviest animal measured in each year? Return the columns year, genus, species_id, and weight.
Joining data
Joining columns: left_join(), inner_join()
Say we have more information about some of our taxa.
count(surveys, taxa)
taxa_iucn <- data.frame(
taxa = c("Bird", "Rabbit", "Rodent"),
iucn = c("NT", "LC", "LC")
)
taxa_iucnLeft join surveys with taxa info by their shared column (the “key”)
surveys_iucn <- left_join(surveys, taxa_iucn, by = "taxa")
head(surveys_iucn)Q10: How many records were there for NT and LC taxa?
left_join() keeps everything from the left. So surveys_iucn has NAs for iucn when taxa wasn’t in taxa_iucn.
Q11: What kind of taxa do the NAs in iucn correspond to?
inner_join() only keeps records where the key is in both tables.
surveys_iucn2 <- inner_join(surveys, taxa_iucn, by = "taxa")Q12: How many rows are in surveys_iucn2? What rows are in surveys_iucn that aren’t in surveys_iucn2?
Wrapping up
Save all your changes to scratch/lesson3.R. Commit and push.
Assessment
For the following assessment, create a Quarto document in reports/ called index.qmd.
Download data
For this assessment, we’ll use the breeding bird phenology from Hällfors et al. (2020a). Their data are on Dryad (Hällfors et al. 2020b). We’ll complement that with the AVONET (Tobias et al. 2022) database of bird functional traits, which is on Figshare.
Download the Dryad dataset. Copy “73_species.csv” and “Traits_73_species.csv” to your data/ folder.
From the AVONET database on Figshare, download “AVONET Supplementary dataset 1.xlsx” do your data/ folder.
Read data
In your Quarto document, add a code chunk that loads the tidyverse and reads the data.
Quarto documents interpret file paths relative to the document, not to your project. Since your Quarto document is in reports/, then data/73_species.csv will point to reports/data/73_species.csv, which doesn’t exist. This is where here::here() function comes in useful! Wrap here::here() around a file path and it’ll fix it to be relative to your project, which is what you want.
This won’t work:
read_csv("data/73_species.csv")
This will work:
read_csv(here::here("data/73_species.csv"))
This also won’t work (notice it’s inverted):
here::here(read_csv("data/73_species.csv"))
Use
read_csv()to read “73_species.csv” and assign it to a variable calledbor_nestlings.Use
read_csv()to read “Traits_73_species.csv” and assign it to a variable calledbor_traits.Use
readxl::read_excel()to read the “AVONET1_BirdLife” sheet from “AVONET Supplementary dataset 1.xlsx” and assign it to a variable calledavonet.
See ?readxl::read_excel for info on how to read a specific sheet from an Excel workbook.
Explore
We’ll explore the boreal bird nestling data together. Follow along in your Quarto document.
Are nestlings showing up earlier in the year over time?
all_birds_trend <- bor_nestlings %>%
group_by(Year) %>%
summarize(mean_doy = mean(Dayofyear))
ggplot(all_birds_trend, aes(Year, mean_doy)) +
geom_point() +
geom_smooth(method = "lm")This may be your first time using ggplot. I’ll give you the ggplot code you need for today. You’ll learn more about it next week.
That combines 73 species. Let’s see the breakdown by species.
species_trends <- bor_nestlings %>%
group_by(Year, Species) %>%
summarize(mean_doy = mean(Dayofyear),
.groups = "drop")
ggplot(species_trends, aes(Year, mean_doy, color = Species)) +
geom_point() +
geom_smooth(method = "lm")So chaotic! What if we just look at the 5 most data-rich species?
data_richness <- bor_nestlings %>%
count(Species)
most_rich <- data_richness %>%
arrange(desc(n)) %>%
slice(1:5)
most_rich_trends <- bor_nestlings %>%
filter(Species %in% most_rich$Species) %>%
group_by(Species, Year) %>%
summarize(mean_doy = mean(Dayofyear),
.groups = "drop")
ggplot(most_rich_trends, aes(Year, mean_doy, color = Species)) +
geom_point() +
geom_smooth(method = "lm")A general pattern, with one conflicting example. What species have the strongest trends?
# I'm giving you a *function* to help here. You'll learn more about those next week.
# Finds the slope of the relationship between y and x
trend <- function(x, y) {
xy_lm <- lm(y ~ x)
coef(xy_lm)[2]
}
# Calculate the trend for all species
bor_trends <- species_trends %>%
group_by(Species) %>%
summarize(doy_trend = trend(Year, mean_doy))Spot check two species
soi <- c("ARDCIN", "LARMIN")
species_trends %>%
filter(Species %in% soi) %>%
ggplot(aes(Year, mean_doy, color = Species)) +
geom_point() +
geom_smooth(method = "lm")Your turn
Find the species with the most positive and most negative trends, then plot just those.
nrow_bor_trends <- nrow(bor_trends) # Use this later
bor_extreme <- bor_trends %>%
# Sort by the day of year trend
___(___) %>%
# Keep just the first (most negative trend) and last (most positive trend) rows
slice(c(___, ___))
# Now plot them
species_trends %>%
filter(Species %in% ___) %>%
ggplot(aes(Year, mean_doy, color = Species)) +
geom_point() +
geom_smooth(method = "lm")Bring in more data
Next we’re going to test the hypothesis that smaller birds have more flexible phenology, i.e. the absolute value of the trend is correlated with smaller body sizes.
To test our hypothesis, we need to add AVONET data to our phenology data by joining. The keys are a bit tricky here!
bor_trends has a column called Species with a 6-letter code.
bor_traits has a column called Abbreviation with the 6-letter code and a column called `Scientific name` with the binomial name.
avonet has column called Species1 with the binomial name.
We need to join bor_trends to bor_traits, then join with avonet.
Welcome to data in the real world!
# First, select and rename important columns
avonet_size <- select(avonet, SciName = Species1, Mass_g = Mass)
bor_sciname <- select(bor_traits,
Species = Abbreviation,
SciName = `Scientific name`)Now join them all together.
bor_trends_size <- bor_trends %>%
left_join(___, by = ___) %>%
left_join(___, by = ___) %>%
mutate(abs_trend = abs(doy_trend))
# Plot it
ggplot(bor_trends_size, aes(Mass_g, abs_trend)) +
geom_point() +
geom_smooth(method = "lm")That’s kind of hard to interpret. Make a new variable, bor_trends_size2, that removes the missing values in Mass_g and keeps only birds smaller than 2000 g with an absolute trend less than 1.
bor_trends_size2 <- ???
ggplot(bor_trends_size2, aes(Mass_g, abs_trend)) +
geom_point() +
geom_smooth(method = "lm")Is our hypothesis supported?
Submission
- Render
index.qmd, which produces a few outputs in yourreports/folder. - Move your output (an HTML file and a folder) from
reports/todocs/. - Enable GitHub pages for your repo.
- Commit your changes and push.
- Open an issue and tag me in it.